Name: Gopinath. S
PATRIC Database
Biological significance of PATRIC Database:
A strategic plan was developed by the National Institute of Allergy and Infectious Diseases (NIAID) for conducting research on Biodefense. They have developed a subsequent watch list of genera from their own defined list of ‘Priority Pathogens’ and they were categorized as A, B and C microbial pathogens with respective priority levels. In order to understand the biology of these priority organisms, to generate new level of diagnostic treatments and vaccines to those to priority microbial pathogens and the scope for all these strategies were outlined and supported by this initiative of NIAID.
In such a way on 2004, for providing the biological scientists with genomic and related data on the above classified list of microbial pathogen and also the transmitting invertebrate vectors, National Institute of Allergy and Infectious Diseases (NIAID) organized and provided funding for eight (BRCs) (BRCs) Bioinformatics Resource Centers. PATRIC and NMPD are two of those eight BRCs, then on the year 2009, these eight bioinformatics Resource Centers are further reorganized through competitive renewal and they were consolidated into four major Bioinformatics Resources Centers through a competitive renewal by NIAID. Then they were consolidated into four BRCs, each one with a distinct resources they all encompass a focus on particular organism: such as eukaryotic pathogens and invertebrate vectors, bacteria and viruses. The influenza virus Resource Database is a single exception, which was initiated to focus specifically on the influenza virus. Likewise, the bacterial BRC was awarded for PATRIC. Then it collaborated with the team of NMPDR in order to produce one of the most wide-ranging resource of data analysis which is available for bacteria. (Alice R. Wattam 2011, 2013)
Solutions provided by the database:
As per the significant reasons behind the creation of PATRIC database, it indeed serves as a unique and a comprehensive resource portal for bacteria by offering vast amount of information in the form of five different data types such as genomic data, Transcriptomics data, Protein-protein interaction, Protein three dimensional structure data, genome Meta data and sequence typing data. In such a way PATRIC provide the user with all possible information that can be provided for the range of microbial pathogens. The major uniqueness of PATRIC database is, unlike other databases it is not just providing the data content but It also provide wide range of tools for each type of data respectively which make it most sophisticated resource portal with data as well as inbuilt tools to obtain and process the data for a particular microbe of interest. As an extension every registered user was provide with a private workspace to import the data directly into the workspace and use any available tools based on the needs and it can be processes and results can analyzed. So PATRIC serves as a significant tool were a user can utilize wide range of options provided in an open source form. (Alice R. Wattam 2011, 2013)
Curation and Validation of the data:
PATRIC collects data and integrates them from various public repositories such as National Centre for Biotechnology Information (NCBI), the Centers for Disease Control (CDC), the USDA United States Department of Agriculture, System Biology for Infectious Disease center and other NIAID-funded projects, such as the studies carried out by individual biological research group or investigators. Some of the data such as Transcriptomics data are manually curated from the repositories but still PATRIC follows different procedures in order to work with different data types from different data providers, they collected data automatically from above mentioned public repositories and most of them are integrated from centers on genes and genomes. Distinct biological entities like genes, DNAs, RNAs, pathways, proteins, and other related data are readily linked to genomes entities.
Every genome data which are available at PATRIC database is annotated using (RAST) Rapid Annotations using Subsystems Technology. Under the NMPDR project RAST is developed as an end-user genome annotation service, which uses a common controlled vocabulary shared with SEED and ModelSEED for supporting the functional content and the inter-comparison of annotations. (Alice R. Wattam 2011, 2013)
Information on Dataset:
Initially PATRIC had data for only eight genera of microbes now it extended greatly to 22 genera with almost 15000 genomes by the end of 2013 and currently it almost crosses 25000, it hosts data from all the NIAID priority pathogenic bacteria, which include 22 genera. A variant of integrated Omics datasets is provide by PATRIC along with the genomes and their annotations

Group I—pathogens newly recognized in the past two decades.
Group II—re-emerging pathogens.
This above table express the summary of descriptions of individual data types and the integrated data at the level of Bacteria and across the target genera is illustrated.
Almost, more than 100 000 bacterial PPIs from various public repositories and around 1300 experimentally characterized host–pathogen protein–protein interactions (PPIs) are incorporated in PATRIC
Sample query search and results:
In the find genomes tab we can use genome browser, scaffold search and genome search options. In find gene, tab we can use BLAST and gene search options. We can various means of pathway search option in the function tab.
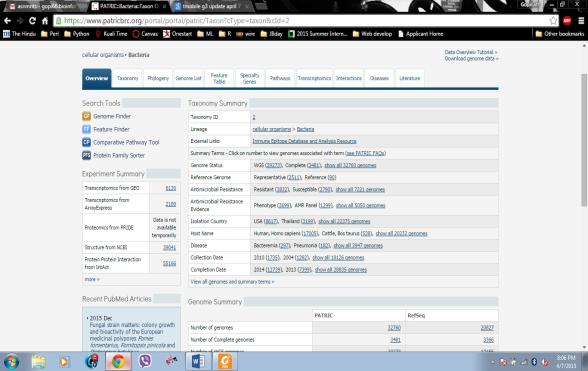
The above figure show the details of all the bacteria and the resources from where the bacterial is collected with the exact statistics.
The below figure shows the complete details of the genomic data present in the PATRIC database and the wide range of tools available for the analysis of the genomic data likewise similar structure or architecture is available for all the other data types in the database.

Similar Tools:
Some of the databases and metagenome project resources which also provide information but not merely equivalent to PATRIC Database.
- IMG/M Database
- Microbial Genome Database for Comparative Analysis EBI Metagenomics
- NCIMB Bacteria Database – CABRI
- Microbial Genome Resource
Drawbacks, Evaluation and comments on the tool:
Generally, PATRIC serves to be one of the most unique dataset resource which can assist researchers who are working on the infectious diseases by providing, wide range of data, tools for analysis and private workspace. Apart from all the positive abilities of the database, The are few drawback which is little uncomfortable for data analysis, Due to large collection of data, the response in loading the results or the query pages is taking lots of time, and It has provide same tools for analyzing different data types in many places. Instead common tools can be provided in one particular tools tab rather providing everywhere which makes the user to spend more time on understanding the architecture of the database also it requires permission in order to access in fully loaded workspace and analysis tools. Apart from that it really does the impressive job. (Evaluation – 4.5/5)
References:
- PATRIC, the bacterial bioinformatics database and analysis resource
Alice R. Wattam1,*, David Abraham1, Oral Dalay1, Terry L. Disz2,3, Timothy Driscoll1, Joseph L. Gabbard1,4, Joseph J. Gillespie5, Roger Gough1, Deborah Hix1, Ronald Kenyon1, Dustin Machi1, Chunhong Mao1, Eric K. Nordberg1, Robert Olson2,3, Ross Overbeek3,6, Gordon D. Pusch6, Maulik Shukla1, Julie Schulman1, Rick L. Stevens2,7, Daniel E. Sullivan1, Veronika Vonstein6, Andrew Warren1, Rebecca Will1, Meredith J.C. Wilson1, Hyun Seung Yoo1, Chengdong Zhang1, Yan Zhang1 and Bruno W. Sobral1,8
- PATRIC: the comprehensive Bacterial Bioinformatics Resource with a focus on Human Pathogenic Species, Infection and Immunity 2011. Alice R. Wattam et al,
- PATRIC: the VBI pathosystems resource integration center. Nucleic Acids Res., 35, D401–D40
Snyder,E., Kampanya,N., Lu,J., Nordberg, E.K., Karur,H., Shukla,M., Soneja,J., Tian,Y.,,T.andYoo,H.(2007)



