After the Bisulfite treatment
5’-ACGTTAGTCG-3’ 5’-ATGTTAGTTG-3’
_
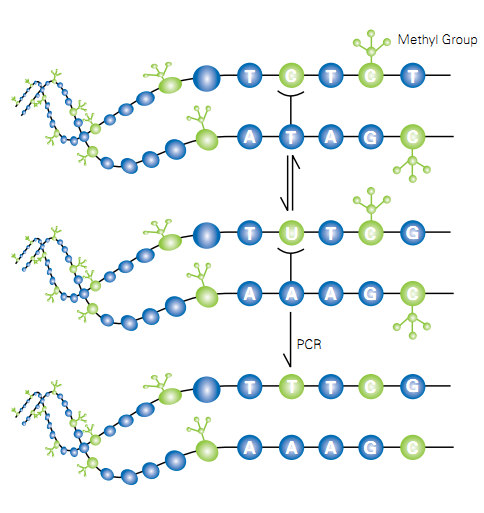
Figure … Bisulfite conversion of unmethylated cytosine followed by PCR. (methyaltion ffpe—11.8.9 –bs con)
Bisulfite-conversion of unmethylated cytocine to uracil is a three step process. First step is the reversible addition of HSO3– to cytosine. Second step is the liberation of NH3 by hydrolysis, and third step is the release of HSO3– to form uracil. The [C-SO3–] formed in first step rapidly regenerates in to [C] because it is unstable in neutral solutions. On the other hand, [C-SO3–] is stable in acid. The equilibrium between [C] and [C-SO3–] is reached rapidly at pH 7 when bisulfite concentration is 0.5M or higher. Second step is the key process of the overall procedure. It is the rate-limiting step for the conversion of [C] to [U-SO3–]. [U-SO3–] is stable in neutral conditions, but can be easily converted to [U] by alkali (Step 3). With respect to pH, the [C] to [U-SO3–] conversion is optimal at pH 5–6. (Hayathsu et al 2008) (Coffee et al 2009)
 _
_
Figure …. Bisulfite conversion of cytosine to uralic
5- methyl cytosine can be deaminated by bisulfite, but the rate is much more slower than cytosine. The rate of deamination is approximately two orders of magnitude lower than that for cytosine. T he deaminated products, thymine-bisulfite adducts are a mixture of two diastereomers. The structures of these isomers are shown in Fig. …..
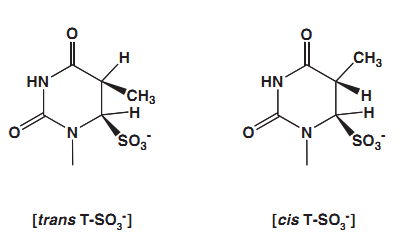
Figure … Two diastereomeric thymine -bisulfite adducts formed in the deamination of 5- methylcytosine.
They are produced in approximately 1 to 1 ratio. The treatment of trans isomer with alkali rapidly generates thymine. In contras the cis adduct, is quite stable. It can be converted to thymine only with a harsh alkaline treatment because cis elimination is difficult. This situation creates an issue in bisulfite modification of methylated cytosine. (Hayathsu et al 2008)
DNA degradation is the major side reaction occurs during this conversion. If the incubation temperature and time of the deamination step is increased DNA degradation will be increased. Therefore bisulfite modification is carried out in 55 C temperature for 3 hours. A total of 500 ng genomic DNA is enough for this modification. (BS 1— BS CONVERSION)
_
1.12.6. Melting curve analysis of combined direct 5’ and 3’ triplet primed PCRs
PCR is performed in the presence of intercalating dyes such as SYBR green 1, eva green and BRYT green. The intercalating dye binds to dsDNA producing fluorescence. When the DNA is heated, the strands begin to separate and the dye is released and fluorescence is reduced. The temperature at which of the 50% DNA is in single stranded is the melting temperature (Tm). Tm is influenced by GC content and length, therefore Tm is an indicative of the identity of a PCR product.
_
Individuals with expanded alleles (PM and FM) from both males and females are produced melt peak profiles that are distinct from those individuals with the NL alleles. Individuals with high GC content have higher Tm than individuals with low GC content.
This approach consists of 2 complementary FMR1 triplet-primed PCR (TP-PCR) assays that use unmodified DNA as template. Both assays are based on the same principal feature use of a primer that anneals within the triplet repeats. The combined analysis of both 5’ and 3’ TP-PCRs avoid false negatives caused by flanking-sequence deletion. This assay works as a rapid first-line screen for FMR1 expansions, thereby reducing the number of samples that require more labor-intensive approaches such as Southern analysis for exact repeat sizing and methylation analysis.
Previously described methods such as conventional PCR and MS PCR are two step procedures, initially PCR and then product analysis. dTP-PCR followed by MCA is a single step assay with high sensitivity and specificity for rapid screening of FXS.
The closed-tube format of this screening strategy eliminates post-PCR transfers, thereby reducing the possibility of human error and sample mix-ups or contamination, an important consideration in the multi-sample screening setting, which is currently minimized via automated sample handling. By eliminating the electrophoresis step, we also reduced the total hands-on and turnaround time.
1.12. 7. Triplet primed PCR and capillary electrophoresis
The CE is the most successful analytical tool for high-throughput, cost-effective, and reliable DNA analysis. CE separates ions based on their electrophoretic mobility with the use of an applied voltage. CE is predominantly used in nucleic acid fragment analysis and DNA sequencing. Triplet-primed repeat PCR combined with CE simplifies detection of FX expanded alleles.
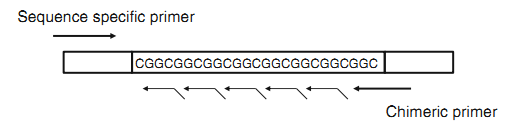
In triplet primed PCR, forward and reverse primer flanks the CGG repeat region. (CGG)5 primer anneal at multiple targets within the repeat region generating multiple amplicons with a length difference of 3 bases. When electrophoresed in agarose, the multiple products appear as a smear. If PCR products subjected to the CE, the smear becomes a characteristic stutter pattern. The number of peaks corresponded to the number of repeat units (tp pcr 2—tppcr 2010). PMs and FMs can be discriminated as expanded from NLs in males and females by evaluating the peak pattern. Then individuals with expanded alleles can be subjected to Southern blot analysis to determine precise allele sizing and methylation pattern. ( Nahhas et al 2012 ). This largely overcome the need for Southern analysis, (Lyon, et al 2010). However CE provides resolution up to approximately 200 repeats. Above 200 all FM alleles migrated late as an aggregate peak independent of length, located at approximately 850 to 1100 bp range () (TPPCR 1—TP PCR 2010)
_
Figure …. Schematic representation of triplet primed PCR
1.12.8. Methyaltion specific triplet primed PCR and capillary elecrophoresis
Another simplified strategy based on fluorescent methylation-specific PCR (ms-PCR) and CE analysis for molecular diagnosis of FXS is available. Sodium bisulfite modification is performed to selectively modify genomic DNA. Then msPCR is perforemed using fluorescently labeled three primer sets with different fluoropores to amplify modified methylated or unmethylated DNA and amplicons are resolved by CE. This assay accurately size the NL and PM in both male and females according to the fluorescent peak sizes and patterns on the electropherogram. (Zhou et al 2006)
1.12. 9. Methyaltion specific triplet primed PCR and Melting curve analysis (msTP-PCR MCA)
In this assay MCA is performed for the amplicons generated from MS PCR. The melting temperature of an amplicon is dependent on its length and base composition. Amplicons generated from methylated templates have 5-methylcytosine in the positions corresponding to cytosine and they have higher melting temperatures than amplicons from unmethylated templates, which contain thymines. When methylation-independent primers are used, this allows the amplification of both methylated and unmethylated sequences which are seen as distinct melting peaks. (Candiloro et al 2011)
Previously described dTP-MCA assay does not discriminate between PMs and FMs. However the msTP-PCR MCA can differentiate PMs from FMs. Therefore only FMs cab be subjected to sizing by Southern analysis. The msTP-PCR MCA assay is rapid and less labor-intensive than Southern blotting or even the newer PCR and capillary electrophoresis– based tests. This assay also ideal for population-based newborn or early childhood screening of FXS. (bs-mca——–ref nus 2013)
1.12.10. Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS)
Southern blot analysis provides the number of the CGG repeats and methylation status for all types of FMR1 alleles. But one of its limitations is that it targets methylation of only a few CpG sites located on the CpG island in 5′ of the repeat region. Recently developed test for FXS targets biomarkers located in the CpG sites.
Two novel epigenetic markers called fragile X–related epigenetic elements 1 and 2 (FREE1 and FREE2) were identified and they are related to FMRP expression (inversely correlated with FMRP production) and cognitive impairment in FM individuals. These elements are located on either side of the repeat region and are different from the sites routinely analyzed using methylation-sensitive SB.
FREE1 is located on the 5‘ region of CGG repeats. FREE2 is located on the exon 1/intron 1 boundary of the FMR1 gene. FREE2 allows identifying FM males and females with 100% specificity and sensitivity but could not distinguish between PM carriers and healthy controls or between PM carriers and high functioning males with un methylated FM alleles (Godler et al 2010, Godler et al 2011, Godler et al 2012, Godler et al 2013, ) .
1.13 Ethical aspects
Genetic testing is a complex ethical and social concern. The use of patient sample requires informed consent from individual. If the study population is children or mentally impaired consent should be obtained from parent or guardian. Explanation of the research and its potential outcome is must. Each individual is entitled to privacy and confidentiality in terms of the protection of their personal data and research findings.
Current guidelines state that genetic testing of children is recommended only if a clear benefit can be offered. Fragile X testing for children less than 18 years of age must be approached carefully, with medical and emotional benefits to the child weighed against potential harms.


 _
_
